- Software
- Open access
- Published:
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes
BMC Genomics volume 13, Article number: 688 (2012)
Abstract
Background
Information about the composition of regulatory regions is of great value for designing experiments to functionally characterize gene expression. The multiplicity of available applications to predict transcription factor binding sites in a particular locus contrasts with the substantial computational expertise that is demanded to manipulate them, which may constitute a potential barrier for the experimental community.
Results
CBS (Conserved regulatory Binding Sites, http://compfly.bio.ub.es/CBS) is a public platform of evolutionarily conserved binding sites and enhancers predicted in multiple Drosophila genomes that is furnished with published chromatin signatures associated to transcriptionally active regions and other experimental sources of information. The rapid access to this novel body of knowledge through a user-friendly web interface enables non-expert users to identify the binding sequences available for any particular gene, transcription factor, or genome region.
Conclusions
The CBS platform is a powerful resource that provides tools for data mining individual sequences and groups of co-expressed genes with epigenomics information to conduct regulatory screenings in Drosophila.
Background
Massive genome-wide characterization projects have dramatically transformed our current view of genes and other elements of the genome [1]. The picture emerging is of a complex regulatory landscape in which multiple actors coincide to perform distinct roles that are fundamental for the appropriate deployment of cellular gene expression programs [2]. Transcription factors (TFs) are protein adaptors that recognize particular regulatory sequences (TF binding sites, TFBSs) in the genome to target the assembly of other protein complexes that ultimately govern gene expression [3]. In fact, precise information about when and where a gene is transcribed is encoded on the sequence and the structure of the genome.
At the sequence level, promoters are regulatory regions located immediately upstream of the gene, which anchor the RNA polymerase transcriptional machinery to the transcription start site (TSS), while enhancers conduct more precise tissue-specific gene expression and can be physically displaced up to hundreds of kilobases from their target. Both promoters and enhancers are non-coding sequences in which multiple TFBSs of about 5 to 15 bp are distributed following a modular organization. Such cis-regulatory modules (CRMs) act as genetic switches and are bound by specific TFs to drive distinct patterns of expression. Comparison of binding landscapes across multiple species have revealed that these functional regulatory regions tend to be highly conserved throughout evolution in many cases [4–6]. The predominant model establishes that direct contact between both enhancers, promoters and TFs, through DNA looping orchestrates the RNA polymerase recruitment to initiate the transcription of the neighbouring gene [7].
At the structural level, chromatin packaging into nucleosomes dynamically shapes the genome, producing a landscape of open and closed regions that can eventually show or mask different pieces of information encoded within the sequence [8]. By interpreting a collection of post-translational modifications of the histone tails at the surface of nucleosomes, chromatin remodeling complexes can force a repositioning of such structural units, resulting in a change in the local conformation of a particular area [9]. Consequentially, modifications in the chromatin structure may confine access of TFs to a subset of regulatory sequences along the genome [10]. Recent studies on epigenomics have unveiled the existence of chromatin signatures that are helpful to distinguish promoters and enhancers from other genome elements [11–13]. Thus, while active gene promoters are in general marked by trimethylation of lys4 of histone H3 (H3K4Me3), distal enhancers are associated with monomethylation (H3K4Me1). However, functional enhancers for active genes exhibit additional acetylation of lys27 of histone H3 (H3K27Ac), while trimethylation of lys27 (H3K27Me3) denotes poised enhancers that are linked to inactive genes [11, 13].
Deciphering the map of regulatory sites and regions that shape the genome is therefore a formidable challenge of major interest, for which computational methods that identify such features can be extremely helpful. Most bioinformatics protocols for regulatory prediction consist in the application of two steps (reviewed in [14–16]): (i) sequence analysis in search of consensus sites derived from catalogs of predictive models or motif discovery approaches; and (ii) evaluation of such predictions, taking into account evolutionary conservation across other species. Recently, additional epigenomic information about histone modification maps has been integrated into other approaches, and this significantly outperforms previous strategies [17–19]. In the last decade, a myriad of bioinformatics solutions have been published that deal with the problem of mapping putative TF sites and predicting regulatory regions (see [20] for a comprehensive listing). As a consequence, scientists must face a plethora of heterogeneous tools in order to characterize a regulatory region, including, among others, genome browsers [21, 22], multiple genome alignments [23, 24], catalogs of functional sites [25–27], software suites of prediction [28, 29], and genome-wide epigenetics profiles [30, 31]. Even though unquestionable progress is observed in this issue, through integrative initiatives such as Galaxy [32], this complex mixture of applications and databases often constitutes an obstacle for basic researchers, who are actually the potential target audience demanding this information. The minimal computational expertise that is required may be prohibitive for many experimentalists, denying them access to this knowledge that could expedite their investigations at the lab bench [33].
Research on transcriptional regulation in Drosophila melanogaster, one of the most intensively studied organisms in biology, is a case in point. In fact, the sequencing of other flies [24] offers a formidable opportunity to decipher the common regulatory circuitry of these species. This information is fundamental when conducting experimental research to elucidate potential relationships between regulators and their targets [34]. More recently, the modENCODE project has released more than 700 genome-wide datasets for dozens of TFs, histone marks, and other regulatory features that promise to drastically push the field of characterization of Drosophila gene regulatory regions forward in the next decade [35]. By and large, fly researchers can work with many resources that provide inestimable access to such information: FlyBase is the major repository of genetic and genomic information on the fruit fly [36], FlyMine is a web platform that integrates external genomics and proteomics resources under the same query interface [37], and modMine provides access to modENCODE data [31]. Specifically for the characterization of TF binding sites, the information is distributed into different resources: FlyTF is a comprehensive catalog of TFs with DNA-binding properties [38], while REDfly, FlyFactorSurvey, and the DNase footprint database are compilations of TFBSs experimentally validated in Drosophila[39–41]. Moreover, Jaspar and Transfac repositories include about 100 predictive models derived from the literature for Drosophila[25, 26].
However, although important efforts are being done to standardize the construction of large-scale collections of regulatory sites [6, 42], it is not trivial for a bench biologist to understand how to deal with this massive volume of information (for examples, see [43, 44]). As a result, there is a need for easy-to-use web integrative resources that perform comparative regulatory analyses on emerging next-generation sequencing data. We present here CBS (Conserved regulatory Binding Sites), an open regulatory platform that offers, under an intuitive graphical interface, a comprehensive map of evolutionarily conserved binding sites and enhancers identified in Drosophila, using a combination of predictive and alignment methods with epigenomic information. Through the introduction of user custom tracks for most popular genome browsers, CBS makes visualization of these regulatory features extremely simple for inexpert users. We demonstrate how CBS can be particularly useful for characterizing functional sequences and conducting in silico regulatory screenings of target genes reported in high-throughput expression experiments.
Implementation
Prediction of conserved TFBSs
CBS integrates regulatory information for 21,984 RefSeq transcripts [45] of 13,678 genes from D. melanogaster (BDGP Release 5/dm3 April 2006). Prior to computational prediction of TFBSs and enhancers, we masked the sequence of all known exons using current gene annotations in RefSeq [46]. We constructed a catalog of 850 predictive models of TFs that integrates 255 weight matrices from Jaspar CORE [25] and 595 weight matrices from Transfac 8.4 [26]. For simplicity, users see the full catalog of matrices organized into 346 distinct TFs, whereby each one may represent occurrences for several matrices that predict the same class of TF. To produce the initial set of predictions, we searched for best occurrences of each predictive matrix on the fruit fly genome with MatScan [47], discarding those hits with a similarity below 85% of each weight matrix. The phastCons program computes conservation scores based on a phylo-HMM with two states (conserved and non-conserved regions) that correspond to the posterior probability that a given alignment column is generated by the conserved state of the phylo-HMM [48]. From the initial pool of predictions, we removed those binding sites within genome regions in the UCSC genome browser [21] that presented on average a probability lower than 0.95 to be conserved across the 12 flies multiz15way alignments [49]. This treatment resulted in a five-fold reduction of binding sites. Users can instruct CBS to include predictions that exhibit a probability higher than 0.95 to be in conserved regions (moderate), or force hits that present a probability of 1 (maximum) to be displayed. All predictions, irrespectively of the conservation score, can be downloaded from CBS website as flat files.
Prediction of putative regulatory enhancers
We gathered ChIP-seq enriched regions for H3K4Me1, H3K4Me3, H3K27Ac, and H3K27Me3, as reported by the modENCODE consortium [35, 50], in the following developmental stages: embryos (0–4 h, 4–8 h, 8–12 h, 12–16 h, 16–20 h, and 20–24 h), larvae (L1, L2, and L3), pupae, and adult males and females (see Additional file 1 for further details). We considered each non-coding genome region that presented a significant H3K4Me1 signal to be a putative enhancer. To separate enhancers from gene promoters, we generated an alternative list of regions exhibiting enrichment in H3K4Me1 that lacked the H3K4Me3 signal. To distinguish active enhancers from poised enhancers, we searched for those genome regions that showed enrichment either in H3K27Ac or H3K27Me3 in the same developmental stage as well (Additional file 2). To focus our search on non-coding sequences (intergenic and intronic regions), we masked previous predictions when overlapping RefSeq exons (Additional file 3). We provide access for graphical display in the CBS website to a subset of these predictions: we discarded those putative enhancers that showed a probability lower than 0.5 to be conserved in 12 flies multiz15way alignments produced by the UCSC Genome Browser [23, 24], removing shorter regions (less than 200 bp) that might produce artifacts from this procedure. Following this protocol over all stages, we ended up with 15,454 putative enhancers (regions with at least H3K4Me1, average size 985.1 bp), 13,326 putative enhancers excluding gene promoters (regions with H3K4Me1 that lack of H3K4Me3, average size 985.3 bp), 6 269 active enhancers (regions with H3K4Me1 and H3K27Ac, average size 936.8 bp) and 4 847 poised enhancers (regions with H3K4Me1 and H3K27Me3, average size 1215.7 bp). In summary, our set of predicted enhancers roughly involves 8% of the fruit fly genome in terms of total coverage.
Data sets from other Drosophila genomes
From available pairwise BLAT alignments between D. melanogaster RefSeq genes and the rest of Drosophilids available in the UCSC genome browser (xenoRefGene tracks), we constructed a putative gene annotation for the following assemblies: D. ananassae (droAna1), D. erecta (droEre1), D. grimshawi (droGri1), D. mojavensis (droMoj2), D. persimilis (droPer1), D. pseudoobscura (dp3), D. sechellia (droSec1), D. simulans (droSim1), D. virilis (droVir2), and D. yakuba (droYak2). In previous works [51, 52], we extracted 1 000 nucleotides upstream of the TSS to define gene promoters. Here we took a more conservative approach and considered the region within 2 000 nucleotides upstream of the corresponding TSS when analyzing multiple promoters for 11 Drosophila genomes. MatScan [47] is used to search for best occurrences of a particular TF on each set of orthologs. The GFF2PS program [53] produces the graphical map of final predictions.
Supporting information
From REDfly v3.2 [39], we gathered 1 830 experimentally validated CRMs, and 1 825 TFBSs reported in the literature. Moreover, we imported the binding instances for 56 TF motifs with conservation confidence values of 60% or higher as identified by Kheradpour et al.[6].
Implementation details
CBS web interface is implemented via a set of PHP scripts to provide access for different MySQL tables that store gene annotations, collections of predictive models, predicted TFBSs/enhancers, and orthologous promoters in multiple species. All these catalogs are publicly distributed as stand-alone flat files at the CBS website. Configurable balloon tooltips introduced in CBS help menus were designed by Sheldon McKay in the Cold Spring Harbor Laboratory for the Generic genome browser [54].
Results and discussion
Database features
Content
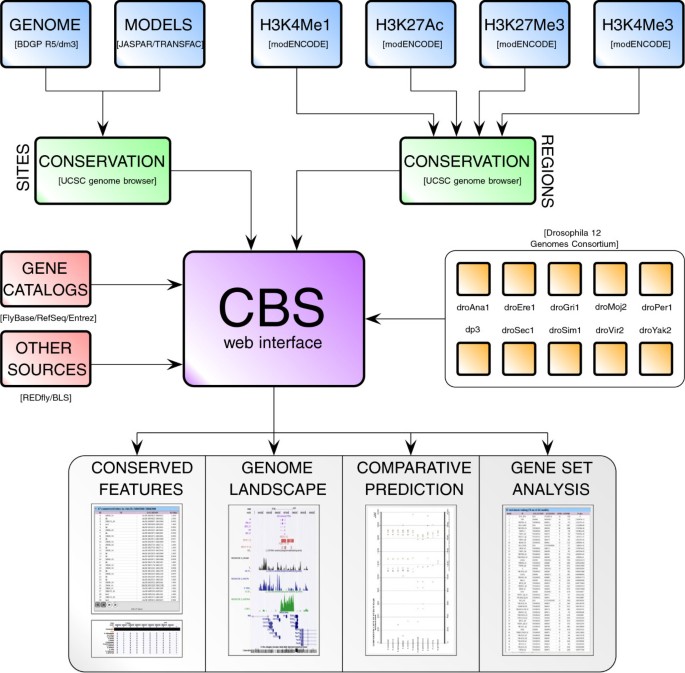
CBS reports a body of predictions and experimental evidence from several sources of information that are combined using a bioinformatics protocol aimed at substantially masking the complexity of such tasks (see Figure 1 and Table 1). For virtually each non-coding region in the fruit fly genome, users can call up the following information (see Implementation): (a) computationally predicted 27,868,614 sites for 346 TFs that are phylogenetically conserved along multiple Drosophilids; (b) 15,454 putative enhancers (6 269 active and 4 847 poised enhancers), computationally inferred from modENCODE ChIP-Seq data [35] throughout all developmental stages; (c) 1 830 CRMs and 1 825 TFBSs gathered from experimental literature in the REDfly database v3.2 [39]; (d) 52,724 computational binding sites predicted by Kheradpour and colleagues [6]; (e) genome-wide ChIP-seq profiles for H3K4Me1, H3K4Me3, H3K27Ac, and H3K27Me3 from modENCODE [35] for all developmental stages; and (f) computational predictions for 346 TFs on 22,763 orthologous promoters of RefSeq genes from 10 Drosophila genomes [21]. CBS functions can be interrogated using multiple equivalent gene name nomenclatures from distinct sources, such as FlyBase, RefSeq, NCBI Entrez, Gene symbol names, and CG codes.
Workflow of the CBS analysis platform. CBS functions can be divided into two categories of evolutionarily conserved information: prediction of regulatory sites, and identification of putative enhancers. The CBS website interface integrates both classes of predictions with other regulatory resources, such as experimentally validated sites and genome-wide profiles of several histone post-transcriptional modifications, in order to characterize in silico a region of the Drosophila genome.
User interface
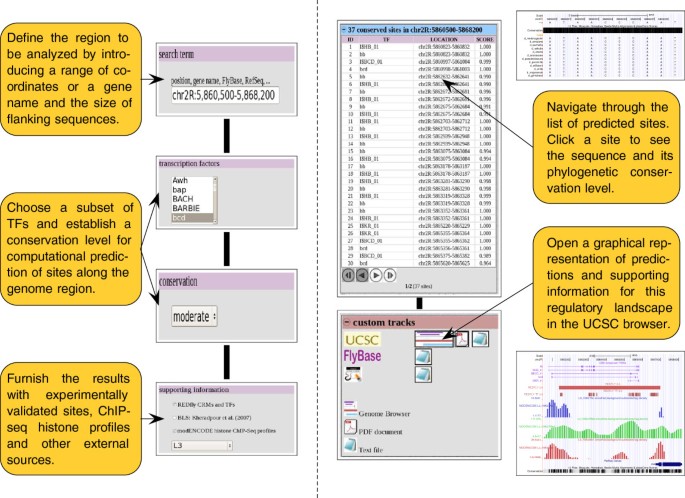
The CBS website is designed to minimize the number of interactive steps that users should follow to end up with the display of the resulting information, offering a common interface to perform each analysis protocol on a particular input set. In consequence, the basic usage of this tool generally requires four fundamental elements (see Figure 2, left): (i) information regarding the locus to be analyzed (gene names or genome coordinates); (ii) class of TFs in which the user is interested; (iii) conservation level between species that is expected for final predictions; and (iv) supporting information from external sources (REDfly [39], BLS [6] and modENCODE histone ChIP-seq profiles [35]) that must be integrated into the final output. Users will find abundant help in the website on the particular options of each CBS function, together with hands-on tutorials that include suggestions about the interpretation of the results.
Basic usage of the CBS interface. Input information that is necessary to interrogate the system in search of predictions and annotations on a particular locus (left). Output provided for the characterization of a genome region in terms of TF binding sites of a certain family (right). Such data is divided into a list of individual sites, and a global representation of these predictions is visualized in a genome browser.
CBS outputs are presented following a similar layout. Thus, results for a query are divided into three main blocks (see Figure 2, right): information about the options selected by the user in previous screens and genes involved in current analysis, the list of individual TFBSs that are predicted in regions of the fruit fly genome that present a significant conservation level among Drosophilids, and the files of custom tracks to produce global representations of the results in several genome browsers as UCSC [21], FlyBase [36], and modENCODE [31]. Knowledge about such resources is graphically presented as independent tracks of annotations that users can easily show or hide, to facilitate the visibility of the final picture [56]. CBS exports the information solicited by the user to different genome browsers, taking advantage of their standard interface. For example, by clicking on the location of a particular site in the list of individual predictions, its sequence can be accessed in the genome using the UCSC genome browser. On the other hand, when it is necessary to visualize the global regulatory landscape of the region that is currently analyzed, users can integrate both predictions and supporting information into a single file following conventional genome browsing standards, by clicking on the graphics icon of this section (see Figure 2, right). As a result, it is feasible to represent TFBSs and enhancers predicted by CBS, together with experimental data and phylogenetic conservation status on the UCSC genome browser framework (Figure 3). A similar graphical representation can be obtained for Gbrowse servers such as FlyBase and modENCODE (see Additional file 4). All these tools permit the incorporation of results from other high-throughput experiments performed by the user, or from external data from the literature, into the final picture for posterior integrative analysis [56].
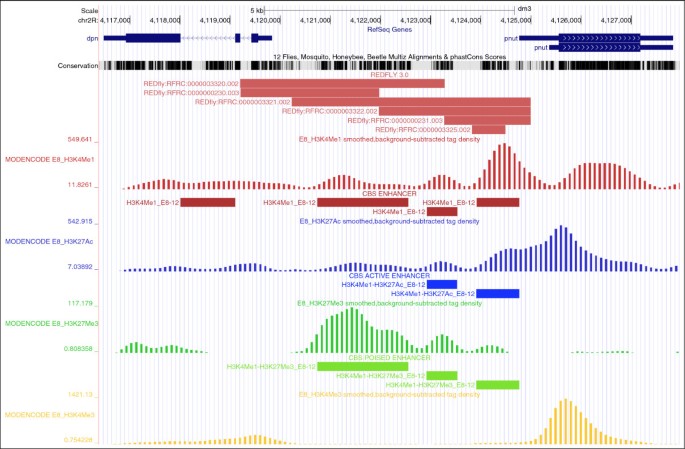
Visualization of CBS information on UCSC genome browser. This example illustrates the graphical display of the global output representation. From top to bottom, the following tracks are shown: RefSeq en gene, UCSC conservation between Drosophila genomes [21], CBS predictions, REDfly experimental CRMs [39], BLS predictions [6], and modENCODE ChIP-seq profiles for chromatin marks associated to enhancer sequences [50]. As external information, we manually incorporated in this case two custom tracks in grey color for Trl and Eve binding profiles as published by modENCODE [50].
Applications
Characterizing genome regions and gene locus
Frequently, wet-lab biologists suspect that a particular TF might participate in the transcriptional regulation of a gene of interest. To address this question, CBS automatically produces the map of predicted TFBSs, demanding a certain conservation level in multiple Drosophilids, and includes epigenetic information into the results to favor the selection of regions that show epigenomics patterns characteristic of enhancers. Therefore, users obtain a list of promising predicted sites at a particular locus together with available experimental information and ChIP-seq profiles for a certain developmental stage. This information can be helpful to discriminate which TFs are feasible as regulators of specific genes, and to find the most solid predictions along the established regulatory region.
As an example, we show the CBS regulatory landscape with predictions for several TFs that are known to be involved in the regulation of the engrailed (en) gene locus (Figure 3). According to REDfly [39], several CRMs have been experimentally reported in the gene promoter and its first intron. We additionally observed enrichment from ChIP-seq results for H3K4Me1, H3K27Ac, and H3K27Me3 from modENCODE [50] in embryos (between 8 h and 12 h). This information can be extremely useful in many occasions to decide which predictions are more reliable. To further highlight the potential of this approach, we manually imported ChIP-seq binding profiles of Trl (GAF) and Eve from embryos (E8—16 h; modENCODE ID 3397 and 3401, respectively) reported by the modENCODE consortium [50] into the same picture (see Figure 3). With this regulatory map, users can accurately decide which potential sites might be more appropriate for validation. In addition, we offer the option to analyze a group of gene promoters to search for the abundance of a particular class of TFs that might explain similar regulatory patterns. For instance, CBS is able to identify within a group of co-regulated genes reported to be relevant for wing imaginal disc regeneration a significant enrichment on AP1 binding sites (see Table 2), which is a downstream target of the JNK signaling pathway that is precisely activated in wound healing stages [51]. Such results confirm the initial regulatory characterization and incorporate additional predictions that enrich the original description.
Exploring putative enhancers
From modENCODE ChIP-seq profiles for several histone marks studied during Drosophila development [50], we built a catalog of potential enhancers —active and poised— that present epigenetics features (see Implementation). For a particular region defined by the user, it is possible to explore the list of CBS predictions of binding sites that are located within potential enhancers in the vicinity of the gene of interest. For example, we show in Figure 4 the upstream promoter region of the dpn gene in which CBS reports the existence of evolutionarily conserved active and poised enhancers using modENCODE ChIP-seq data for embryos (8–12 h) that overlap with REDfly experimental annotations. This set of CBS enhancers is freely available from the website and can be used to annotate a particular genome region in order to explore its transcriptional regulation patterns (Additional file 5). To teach readers about the potential of such predictions, we have exemplified them in two scenarios:
Prediction of putative enhancers in the fruit fly genome with CBS. Here we show the dpn gene locus in which REDfly annotated six CRMs (shown in red); CBS reported several active and poised enhancers (shown in blue and green, respectively) that overlap with the experimental annotations from 8–12 h–old embryos, according to modENCODE chromatin profiles.
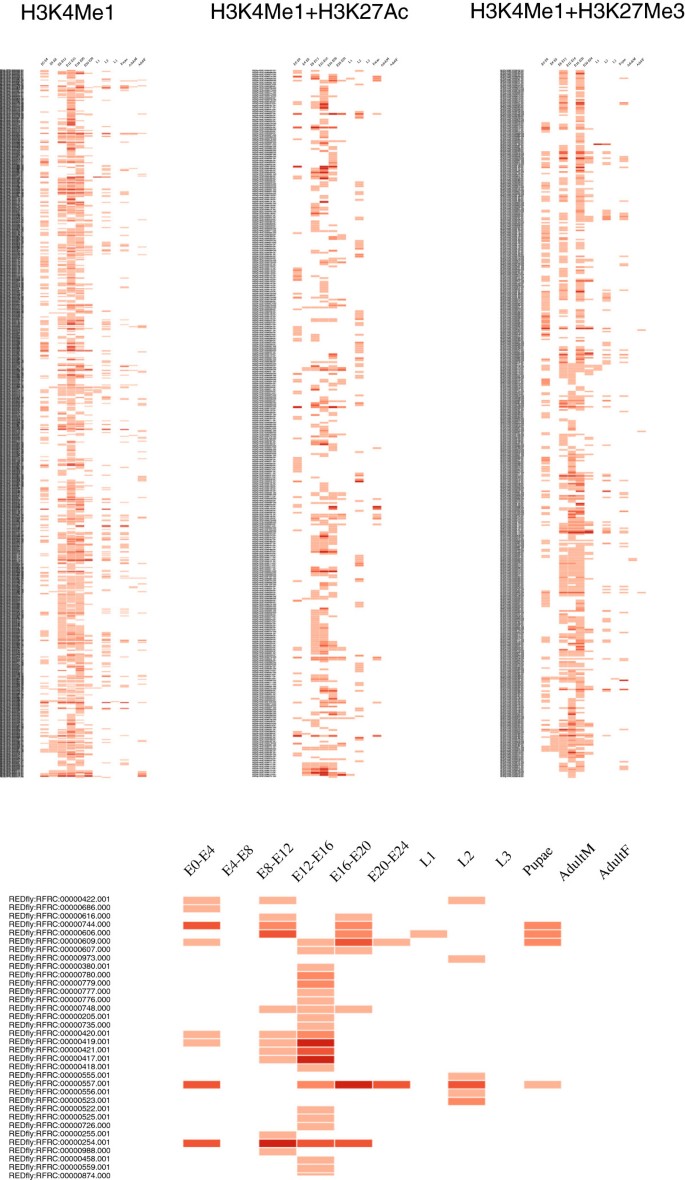
(a) Epigenomics characterization of a set of regulatory sequences. CBS putative enhancers can be extremely useful for studying changes in the activation of regulatory sequences throughout development. Thus, users can take advantage of this information to virtually reproduce these patterns on a particular data set. In Figure 5, we propose to explore how the full collection of REDfly CRMs [39] exhibit different epigenomics features along distinct developmental stages. Here, we have graphically represented this information on a heatmap in which the presence/absence of any of the chromatin signatures available in CBS for each individual regulatory region is denoted in red. To study which CRMs present equivalent regulatory patterns, posterior clustering analysis might be introduced on the resulting pictures. We noticed that for these sequences, such epigenomics signals are likewise more abundant in embryonic stages, which fits well with the fact that REDfly is biased towards embryonic enhancers (because most experiments in the literature are focused on that developmental stage). To study in detail this issue, we analyzed H3K4Me1, H3K27Ac, and H3K27Me3 ChIP-seq–enriched regions from modENCODE stratified by developmental state, in which a similar bias is not expected. In all the scenarios we studied (individual marks, combination of marks, and non-exonic filtering of ChIP regions), we consistently found that H3K4Me1 significantly peaked at embryonic stages (Figure 6). This indicates that most regulatory activity takes place at first developmental stages as previously suggested [57].
A heatmap of REDfly CRMs annotated with the catalog of putative enhancers provided in CBS. At the top are the chromatin signatures of REDfly entries [39] that can be associated with CBS H3K4Me1 enhancers, active enhancers, and poised enhancers, respectively. At the bottom, a magnification of the active enhancer map is shown to illustrate the distinct patterns of developmental stages for a fraction of these CRMs. The darker the intensity of the red color, the higher the number of putative enhancers overlapping a particular CRM. Please note that CBS does not directly produce this type of representation.
Genome coverage of each combination of modENCODE histone profiles to identify putative enhancers throughout all developmental stages of the fruit fly. (First pannel) Distribution of chromatin signatures as provided by modENCODE [50]. (Middle pannel) Distribution of H3K4Me1 and each intersection with a second histone mark. (Last pannel) Distribution of H3K4Me1 and each intersection with a second histone mark when hits overlapping with RefSeq exons were removed.
(b) Identification of putative enhancers in gene-free regions on the fruit fly genome. We consider that our set of predictions can assist in the characterization of novel regulatory regions. Gene-free regions in Drosophila genomes are a case in point. We have detected up to 48 regions of at least 50 Kbp in the fruit fly genome that lack RefSeq transcripts. We characterized the chromatin signatures of these in order to explore putative enhancers. In 20 of the 48 regions (42%, P value < 10-5), we were able to identify at least one evolutionarily conserved enhancer (see Table 3). In most cases, these regions do not exhibit enrichments of the characteristic H3K4Me3 promoter mark, confirming the absence of active genes within these sequences. In addition, it was possible to report changes in the same predictions between different developmental stages as well (Additional file 6). The annotation of such novel elements might be refined with the mapping of CBS predictions of TFBSs within each one of these regions.
Performing comparative genomics
Genome sequencing of multiple Drosophila species finished in 2007 provided a huge volume of data that still remains to be explored [24], and important efforts are conducted to improve the annotation of such genomes [31, 36]. Comparative analysis using D. melanogaster, for which more accurate information is available, can be indeed very effective. A precise sequence comparison method is necessary for the success of phylogenetic footprinting searches in gene regulatory sequences [58]. It is possible, though, that information about the sequence is scarce or even corrupted in certain regions of the genome, which affects the quality of the final alignment. To prevent these problems and provide an alternative comparative method, we have constructed a compilation of orthologous promoters in all species. Therefore, CBS is able to elaborate simultaneously the map of predictions along the promoter of a particular gene in up to 11 Drosophila genomes (see Implementation).
To show the utility of this tool, we have focused the analysis on the promoter of the E(spl) gene. During Drosophila development, the Notch signaling pathway through Su(H) is thought to upregulate the expression of E(spl). When analyzing this sequence in 11 Drosophila genomes, CBS uncovered two regulatory modules constituted of Su(H) + E-box sites that are conserved along the gene promoter (see Figure 7). The first module, located around the positions 1 600–1 800, is conserved in all species, in accordance with prior publications [59]. On the other hand, we found a novel second cluster of sites upstream of the first one, which is partially conserved (note that D. mojavensis, D virilis, and D. grimshawi do not show the same arrangement). Although further work is necessary to solidify this prediction, we believe this example illustrates how CBS can be useful in reconstructing the regulatory signature of other genes in these species.
Comparative map of predictions along the promoter of E(spl) in 11 species of Drosophila. TSS is annotated at position 2 000 of each sequence. Predicted sites for Su(H) are depicted in yellow, and predicted sites for E-box motifs are indicated in blue. Two putative regulatory modules containing the Su(H) + E-box composite are denoted with a black square. The phylogenetic tree was manually incorporated into the picture provided by CBS to show the evolutionary distance among species.
Quality assessment of predictions
We initiated a procedure of evaluation of the quality of CBS predictions based on the study of how variations in the conservation level of sequences may affect the accuracy of CBS predictions. However, it is important to take into account multiple factors that can influence a potential assessment of such data: (i) experimental evidence may be relevant only for a particular developmental stage or tissue, while most promising computational predictions might actually reconstruct the full map of binding affinities in multiple scenarios; (ii) the quality of the predictive models introduced into this analysis may affect the amount of false positives, which will be different for each TF; (iii) published consensus sequences for TFs are constructed for a few examples, while high-throughput experiments can uncover novel sequences that will not be recognized using current predictive models; and (iv) whether a genome region is enriched in a ChIP-seq signal or not in comparison to a certain control is decided by peak-calling software, which can be configured to be more or less strict, producing variations in the sets of annotated binding regions to be used in the evaluation.
For the aforementioned reasons, we consider that a general assessment of multiple TFs using experimental data is beyond the scope of this work. Thus, we focused our study on particular modENCODE ChIP-seq binding regions for several TFs [50] – Trl (E16–24 h, modENCODE ID 3238), h (E0–8 h, modENCODE ID 2574), ttk (E0–12 h, modENCODE ID 615), sens (E4–8 h, modENCODE ID 2577) – and in modENCODE ChIP-seq hotspots (loci with higher levels of TF binding activity). Sequence conservation along ChIP-seq binding sites of each TF presents heterogeneous patterns, ranging from 0.63 (sens) to 0.34 (ttk) on average (see Additional file 7). In this scenario, we decided to study in detail one of these TFs (GAF/Trl) to learn about conservation and quality of predictions. In this case, when plotting the distribution of Trl true positive real binding sites against the total number of computational predictions, the curve reaches the maximum deviation from the random distribution between 0.40–0.60 (see Additional file 8, top). However, it is interesting to mention that when computing the ratio between the total number of predictions along different conservation levels and the number of successfully identified ChIP-seq binding sites, we observed better predictive values for higher sequence conservation (see Additional file 8, bottom). Nonetheless, taking into account our previous considerations, we would like to emphasize that these results cannot necessarily be extrapolated to other TFs in other developmental stages. Although it is important to capture each class of evidence for each TF and gene regulatory region, we offer only those binding sites that show strong evolutionarily conservation in the CBS website to avoid excessive exposition of information. Those predictions normally fit better with experimentally validated sites (see matches between CBS predictions and published binding sites on even-skipped gene stripe 2 enhancer in Additional file 9). For further analysis of the whole body of predictions, we recommend that more expert users download the complete set of predictions for all TFs along the fruit fly genome, which is available as standard GFF files from the CBS website.
To evaluate the accuracy of enhancer predictions, we took into account the collection of 1 830 experimentally validated CRMs published in REDfly v3.2 [39]. To circumvent the overlap between REDfly annotations along the genome, we merged coincident sequences to produce a data set of 726 non-overlapping regulatory regions. When mapping the relationship on the fruit fly genome between merged REDfly CRMs and our set of predictions in any developmental stage, we obtained the following results (P value is 0 in all cases): 597 CRMs (82%) present putative enhancer marks (at least H3K4Me1), 357 CRMs (49%) present active enhancer marks (H3K4Me1 and H3K27Ac), and 325 CRMs (45%) present poised enhancer marks (H3K4Me1 and H3K27Me3). In order to establish the importance of sequence conservation, we repeated the assessment using only those predicted enhancers of each class that exhibit higher sequence conservation (UCSC multiz15way alignments with an average score of 0.50 or more), which improved previous results in most cases (P value is significant in all cases): 424 of 597 CRMs (71%) confirmed by putative enhancers, 227 of 357 CRMs (64%) confirmed by active enhancers, and 248 of 325 CRMs (76%) confirmed by poised enhancers. In contrast, only 92 CRMs (13%) were associated to putative enhancers when this search was reproduced with predictions that exhibit weak sequence conservation among Drosophila genomes.
Comparison with other tools
A quick exploration of recent literature in search of computational tools to annotate regulatory regions can produce dozens of positive hits. In fact, a deluge of effective bioinformatics approaches have been published that aim to make the computational prediction of TFBSs easier for non-expert users (see for further review [20, 60, 61]). Although it is beyond the scope of this work to evaluate the performance of each predictor on standard data sets (see [62] and [63] for exhaustive evaluation on motif finding and promoter identification, respectively), we have annotated several features of the most popular tools as compared to our approach. From the different attributes that characterize these applications, we will focus our discussion on the following ones: (i) availability of epigenomics information; (ii) evaluation of predictions with phylogenetic footprinting data; (iii) classes of sequences that can be studied; and (iv) prediction through a web service (see Table 4 for the full list of applications and features).
It is likely that current predictors will include chromatin and epigenomics information in the near future to refine their predictive output; however, to our knowledge, the CBS platform of regulatory predictions is the first tool that integrates information on computationally identified TFBSs and histone modification marks to characterize Drosophila genomes. On the other hand, with the rapid sequencing of multiple genomes, a significant number of studies that characterize regulatory regions have introduced the evaluation of conservation across species as another discriminant factor to get rid of false positives. Although it is assumed that this information is valuable for this process in most scenarios, just a handful of currently available tools take advantage of this option to reduce the size of the set of predictions (CBS, Centipede [18], BLS [6], GenomeSurveyor [68], see Table 4).
It is assumed currently that not only promoter sequences but also distal enhancers and introns of genes may harbor functional binding sites. However, we still observed a bias in the number of applications exclusively implemented towards the characterization of promoter sequences (e.g. TFMExplorer [65], DoOPSearch [66], COTRASIF [67], Mapper [69], CoreTF [70], and Pscan [71], see Table 4). For this reason, tools such as CBS or GenomeSurveyor [68], which allow users to conduct virtual screenings on the full sequence of genomes, are much more informative. Finally, it is important to take into account the effort that non-expert users need to make to obtain the final results for a particular query. Although many approaches provide access to their maps of predictions through web servers that intuitively accept queries to output appropriate results (e.g. TFMExplorer [65], Mapper [69], and Pscan [71]), only CBS is able to automatically exchange information with popular genome browsers to reconstruct global prediction maps along the genome from custom tracks.
Limitations of CBS predictions
Considering that informatics predictions offer information of limited value in certain situations, we would like to stress potential pitfalls that can affect the quality of CBS predictions: (i) poor specificity of published predictive models for some TFs due to the lack of biological information; (ii) uncompleteness of genome assemblies and the effect on the resulting inter-species alignments; (iii) strong sequence conservation sites that are not always functional because other elements, such as chromatin structure, can influence their activity; (iv) functional sites that are species-specific can be omitted when performing phylogenetic footprinting; (v) ChIP-seq experiments performed on heterogeneous cell populations might produce contradictory epigenomic profiles as a result of the overlap between distinct regulatory landscapes from different tissues; (vi) software for detection of regions enriched in ChIP-seq signals relies on the application of variable thresholds, which affects the final set of results on each case; and (vii) existence of alternative promoters and alternatively spliced forms that can harbor functional sites specific on certain isoforms. Taking into account these considerations and proceeding with caution in every situation, we believe that CBS predictions will constitute a highly valuable resource for researchers.
Future development
Emerging high-throughput technologies are rapidly changing the class of regulatory information that is available to perform computational analysis of genomes. To appropriately evolve into this scenario, we are working on multiple lines of research to update CBS in the future: (a) incorporate published ChIP-seq profiles of TFs in Drosophila to evaluate the consistency of computational predictions; (b) include profiles for other histone marks that are thought to participate in gene transcription regulation (e.g. H3K9Ac); (c) integrate ChIP data and computational maps of Polycomb and Trithorax binding sites, which constitute the core of the regulatory machinery for many genes throughout the fly development; (d) display RNA-seq from modENCODE [77] on each developmental stage to inform about the expression of genes; (e) include experimental information on other Drosophila genomes when available; (f) integrate tools such as TF-map alignments [47] or EEL [72] to provide maps that harmonize predictions and experimental data; and (g) implement a mechanism to perform automatical updates of external repositories integrated in CBS.
Conclusions
The CBS platform is an open resource developed to bridge the gap between experimental researchers and computational predictive methods. Access to this information is provided through a friendly and intuitive web interface, allowing users to easily gain knowledge. Importantly, flexibility of use in CBS does not require a limitation in the volume of information provided to users. In fact, we offer here the most comprehensive compilation of phylogenetically conserved binding sites and epigenomics predictions in the fruit fly genome published to date. In summary, we believe that CBS constitutes an excellent tool for assisting the experimental characterization of regulatory regions of Drosophila.
Availability and requirements
Project name: CBS
Project home page: http://compfly.bio.ub.es/CBS/
Operating system(s): Platform independent
Programming language: PHP
License: Free
Any restrictions to use by non-academics: None
Abbreviations
- CBS:
-
Conserved binding sites
- TF:
-
Transcription factor
- TFBS:
-
TF binding site
- CRM:
-
Cis-regulatory module
- modENCODE:
-
Model organism encyclopedia of DNA elements
- ChIP-seq:
-
Chromatin immunoprecipitation followed by massive DNA sequencing
- H3K4Me1:
-
Monomethylation of lys4 of histone H3
- H3K27Ac:
-
Acetylation of lys27 of histone H3
- H3K27Me3:
-
Trimethylation of lys27
- H3K4Me3:
-
Trimethylation of lys4 of histone H3.
References
Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M: What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007, 17 (6): 669-681. 10.1101/gr.6339607.
Splinter E, de Laat W: The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J. 2011, 30 (21): 4345-4355. 10.1038/emboj.2011.344.
Zaret KS, Carroll JS: Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011, 25 (21): 2227-2241. 10.1101/gad.176826.111.
He Q, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J: High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet. 2011, 43 (5): 414-420. 10.1038/ng.808.
Borok MJ, Tran DA, Ho MC, Drewell RA: Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila. Development. 2010, 137 (1): 5-13. 10.1242/dev.036160.
Kheradpour P, Stark A, Roy S, Kellis M: Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007, 17 (12): 1919-1931. 10.1101/gr.7090407.
Sakabe NJ, Savic D, Nobrega MA: Transcriptional enhancers in development and disease. Genome Biol. 2012, 13 (1): 238-10.1186/gb-2012-13-1-238.
Jiang C, Pugh BF: Nucleosome positioning and gene regulation: advances through genomics. Nature reviews. 2009, 10 (3): 161-172.
Kouzarides T: Chromatin modifications and their function. Cell. 2007, 128 (4): 693-705. 10.1016/j.cell.2007.02.005.
Li B, Carey M, Workman JL: The role of chromatin during transcription. Cell. 2007, 128 (4): 707-719. 10.1016/j.cell.2007.01.015.
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al: Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010, 107 (50): 21931-21936. 10.1073/pnas.1016071107.
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al: Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007, 39 (3): 311-318. 10.1038/ng1966.
Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J: A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010, 470 (7333): 279-283.
Hannenhalli S: Eukaryotic transcription factor binding sites--modeling and integrative search methods. Bioinformatics (Oxford, England). 2008, 24 (11): 1325-1331. 10.1093/bioinformatics/btn198.
Vingron M, Brazma A, Coulson R, van Helden J, Manke T, Palin K, Sand O, Ukkonen E: Integrating sequence, evolution and functional genomics in regulatory genomics. Genome Biol. 2009, 10 (1): 202-10.1186/gb-2009-10-1-202.
Wasserman WW, Sandelin A: Applied bioinformatics for the identification of regulatory elements. Nature reviews. 2004, 5 (4): 276-287. 10.1038/nrg1315.
Ernst J, Plasterer HL, Simon I, Bar-Joseph Z: Integrating multiple evidence sources to predict transcription factor binding in the human genome. Genome Res. 2010, 20 (4): 526-536. 10.1101/gr.096305.109.
Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK: Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011, 21 (3): 447-455. 10.1101/gr.112623.110.
Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE: Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2011, 44 (2): 148-156.
Van Loo P, Marynen P: Computational methods for the detection of cis-regulatory modules. Brief Bioinform. 2009, 10 (5): 509-524. 10.1093/bib/bbp025.
Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al: The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012, 40 (Database issue): D918-D923.
Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al: Ensembl 2012. Nucleic Acids Res. 2012, 40 (Database issue): D84-D90.
Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al: 28-way vertebrate alignment and conservation track in the UCSC genome browser. Genome Res. 2007, 17 (12): 1797-1808. 10.1101/gr.6761107.
Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al: Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007, 450 (7167): 203-218. 10.1038/nature06341.
Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A: JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010, 38 (Database issue): D105-D110.
Wingender E: The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008, 9 (4): 326-332. 10.1093/bib/bbn016.
Blanco E, Farre D, Alba MM, Messeguer X, Guigo R: ABS: a database of annotated regulatory binding sites from orthologous promoters. Nucleic Acids Res. 2006, 34 (Database issue): D63-D67.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS: MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Web Server issue): W202-W208.
Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J: RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011, 39 (Web Server issue): W86-W91.
Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al: ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012, 40 (Database issue): D912-D917.
Contrino S, Smith RN, Butano D, Carr A, Hu F, Lyne R, Rutherford K, Kalderimis A, Sullivan J, Carbon S, et al: modMine: flexible access to modENCODE data. Nucleic Acids Res. 2012, 40 (Database issue): D1082-D1088.
Goecks J, Nekrutenko A, Taylor J: Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11 (8): R86-10.1186/gb-2010-11-8-r86.
Kumar S, Dudley J: Bioinformatics software for biologists in the genomics era. Bioinformatics (Oxford, England). 2007, 23 (14): 1713-1717. 10.1093/bioinformatics/btm239.
Elnitski L, Jin VX, Farnham PJ, Jones SJ: Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006, 16 (12): 1455-1464. 10.1101/gr.4140006.
Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al: Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science (New York, NY. 2010, 330 (6012): 1787-1797.
McQuilton P, Pierre SE, Thurmond J: FlyBase 101 - the basics of navigating FlyBase. Nucleic Acids Res. 2012, 40 (D1): D706-D714. 10.1093/nar/gkr1030.
Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, et al: FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007, 8 (7): R129-10.1186/gb-2007-8-7-r129.
Pfreundt U, James DP, Tweedie S, Wilson D, Teichmann SA, Adryan B: FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Res. 2010, 38 (Database issue): D443-D447.
Gallo SM, Gerrard DT, Miner D, Simich M, Des Soye B, Bergman CM, Halfon MS: REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 2010, 39 (Database issue): D118-D123.
Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, Basciotta MD, Brasefield JA, Zhu C, Asriyan Y, Lapointe DS, et al: FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 2011, 39 (Database issue): D111-D117.
Bergman CM, Carlson JW, Celniker SE: Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics (Oxford, England). 2005, 21 (8): 1747-1749. 10.1093/bioinformatics/bti173.
Down TA, Bergman CM, Su J, Hubbard TJ: Large-scale discovery of promoter motifs in Drosophila melanogaster. PLoS Comput Biol. 2007, 3 (1): e7-10.1371/journal.pcbi.0030007.
Nardone J, Lee DU, Ansel KM, Rao A: Bioinformatics for the 'bench biologist': how to find regulatory regions in genomic DNA. Nat Immunol. 2004, 5 (8): 768-774. 10.1038/ni0804-768.
Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J: Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc. 2008, 3 (10): 1578-1588. 10.1038/nprot.2008.97.
Pruitt KD, Tatusova T, Brown GR, Maglott DR: NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012, 40 (Database issue): D130-D135.
Pruitt KD, Tatusova T, Maglott DR: NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35 (Database issue): D61-D65.
Blanco E, Messeguer X, Smith TF, Guigo R: Transcription factor map alignment of promoter regions. PLoS Comput Biol. 2006, 2 (5): e49-10.1371/journal.pcbi.0020049.
Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al: Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15 (8): 1034-1050. 10.1101/gr.3715005.
Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al: Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007, 450 (7167): 219-232. 10.1038/nature06340.
Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al: A cis-regulatory map of the Drosophila genome. Nature. 2011, 471 (7339): 527-531. 10.1038/nature09990.
Blanco E, Ruiz-Romero M, Beltran S, Bosch M, Punset A, Serras F, Corominas M: Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev Biol. 2010, 10: 94-10.1186/1471-213X-10-94.
Perez-Lluch S, Blanco E, Carbonell A, Raha D, Snyder M, Serras F, Corominas M: Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res. 2011, 39 (11): 4628-4639. 10.1093/nar/gkq1322.
Abril JF, Guigo R: gff2ps: visualizing genomic annotations. Bioinformatics (Oxford, England). 2000, 16 (8): 743-744. 10.1093/bioinformatics/16.8.743.
Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al: The generic genome browser: a building block for a model organism system database. Genome Res. 2002, 12 (10): 1599-1610. 10.1101/gr.403602.
Maglott D, Ostell J, Pruitt KD, Tatusova T: Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011, 39 (Database issue): D52-D57.
Cline MS, Kent WJ: Understanding genome browsing. Nat Biotechnol. 2009, 27 (2): 153-155. 10.1038/nbt0209-153.
Bogdanovic O, van Heeringen SJ, Veenstra GJ: The epigenome in early vertebrate development. Genesis. 2012, 50 (3): 192-206. 10.1002/dvg.20831.
Ureta-Vidal A, Ettwiller L, Birney E: Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nature reviews. 2003, 4 (4): 251-262.
Maeder ML, Polansky BJ, Robson BE, Eastman DA: Phylogenetic footprinting analysis in the upstream regulatory regions of the Drosophila enhancer of split genes. Genetics. 2007, 177 (3): 1377-1394. 10.1534/genetics.107.070425.
Broos S, Hulpiau P, Galle J, Hooghe B, Van Roy F, De Bleser P: ConTra v2: a tool to identify transcription factor binding sites across species, update 2011. Nucleic Acids Res. 2011, 39 (Web Server issue): W74-W78.
Narlikar L, Ovcharenko I: Identifying regulatory elements in eukaryotic genomes. Brief Funct Genomic Proteomic. 2009, 8 (4): 215-230. 10.1093/bfgp/elp014.
Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al: Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005, 23 (1): 137-144. 10.1038/nbt1053.
Abeel T, Van de Peer Y, Saeys Y: Toward a gold standard for promoter prediction evaluation. Bioinformatics (Oxford, England). 2009, 25 (12): i313-320. 10.1093/bioinformatics/btp191.
Won KJ, Ren B, Wang W: Genome-wide prediction of transcription factor binding sites using an integrated model. Genome Biol. 2010, 11 (1): R7-10.1186/gb-2010-11-1-r7.
Tonon L, Touzet H, Varre JS: TFM-Explorer: mining cis-regulatory regions in genomes. Nucleic Acids Res. 2010, 38 (Web Server issue): W286-W292.
Sebestyen E, Nagy T, Suhai S, Barta E: DoOPSearch: a web-based tool for finding and analysing common conserved motifs in the promoter regions of different chordate and plant genes. BMC Bioinformatics. 2009, 10 (Suppl 6): S6-10.1186/1471-2105-10-S6-S6.
Tokovenko B, Golda R, Protas O, Obolenskaya M, El'skaya A: COTRASIF: conservation-aided transcription-factor-binding site finder. Nucleic Acids Res. 2009, 37 (7): e49-10.1093/nar/gkp084.
Kazemian M, Brodsky MH, Sinha S: Genome Surveyor 2.0: cis-regulatory analysis in Drosophila. Nucleic Acids Res. 2011, 39 (Web Server issue): W79-W85.
Riva A: The MAPPER2 Database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2012, 40 (Database issue): D155-D161.
Hestand MS, van Galen M, Villerius MP, van Ommen GJ, den Dunnen JT, Hoen PA: CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes. BMC Bioinformatics. 2008, 9: 495-10.1186/1471-2105-9-495.
Zambelli F, Pesole G, Pavesi G: Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009, 37 (Web Server issue): W247-W252.
Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J: Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006, 124 (1): 47-59. 10.1016/j.cell.2005.10.042.
Gonzalez S, Montserrat-Sentis B, Sanchez F, Puiggros M, Blanco E, Ramirez A, Torrents D: ReLA, a local alignment search tool for the identification of distal and proximal gene regulatory regions and their conserved transcription factor binding sites. Bioinformatics (Oxford, England). 2012, 28 (6): 763-770. 10.1093/bioinformatics/bts024.
Blanchette M, Tompa M: FootPrinter: a program designed for phylogenetic footprinting. Nucleic Acids Res. 2003, 31 (13): 3840-3842. 10.1093/nar/gkg606.
Berezikov E, Guryev V, Plasterk RH, Cuppen E: CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting. Genome Res. 2004, 14 (1): 170-178.
Aerts S, Van Loo P, Thijs G, Mayer H, de Martin R, Moreau Y, De Moor B: TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005, 33 (Web Server issue): W393-W396.
Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al: The developmental transcriptome of Drosophila melanogaster. Nature. 2011, 471 (7339): 473-479. 10.1038/nature09715.
Ludwig MZ, Patel NH, Kreitman M: Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998, 125 (5): 949-958.
Acknowledgements
We sincerely thank all the researchers involved in multiple international consortiums of genome sequencing, annotation, and visualization for granting open access of such resources to the scientific community. We are grateful to Josep F. Abril, Sílvia Pérez-Lluch, Ada Repiso, Natalia Mora, and Marina Ruiz-Romero for helpful discussions on the web server and the manuscript. E. Blanco and M. Corominas were supported by the Bioinformatics Platform of CSD2007-00008 from Ministerio de Ciencia e Innovación (MICINN), Spain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EB and MC conceived the bioinformatics analysis. EB performed the bioinformatics experiments. EB and MC wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
12864_2012_4702_MOESM1_ESM.pdf
Additional file 1: List of modENCODE histone modification profiles along each developmental stage that are incorporated into the prediction of CBS enhancers. For each ChIP-seq profile, the following information is given: histone mark, developmental stage, number of regions significantly enriched on this sample as compared to a control as reported by modENCODE, genome coverage, and the NCBI-GEO accession code. Additional file 2. List of combinations between modENCODE histone modification profiles along each developmental stage that are incorporated into the prediction of CBS enhancers. For each combination we show this information: combination of histone marks, developmental stage, genome coverage and percentage of H3K4Me1 regions that present intersection with the second mark. Additional file 3. List of combinations between modENCODE histone modification profiles along each developmental stage that are incorporated into the prediction of CBS enhancers after removing those regions overlapping RefSeq exons. For each combination, the set of histone marks, developmental stage, and genome coverage is given. Additional file 4. Visualization of CBS information on the modENCODE genome Browser. The following information is displayed (from top to bottom): FlyBase en gene annotation CBS predictions for several TFs that are known to participate in the regulation of this gene, REDfly experimental CRMs on this locus, BLS predictions, and ChIP-seq information about H3K4Me1, H3K27Ac, and H3K27Me3, as provided by modENCODE. Additional file 5. Dynamic regulatory pattern landscape along a genome region in embryos (12–16 h). The following information tracks are displayed along this fragment of 300 kb: (i) modENCODE H3K4Me1 ChIP-seq profile in red, H3K27Ac in blue, and H3K27Me3 in green; (ii) CBS evolutionarily conserved enhancers derived from previous profiles; active enhancers are highlighted in blue, and poised enhancers, in green; and (iii) RefSeq gene annotation and UCSC conservation tracks. Additional file 6. Characterizing putative enhancers in a gene-free region with CBS. A region of 50 kb in chromosome 2L that does not contain any RefSeq annotation is shown. The H3K4Me1/H3K4Me3 profiles, and the set of putative enhancers evolutionarily conserved predicted by CBS at this locus, are shown for embryos of 0–4 h and 12–16 h. Additional file 7. Conservation levels in TF binding regions of five different ChIP-seq experiments from the modENCODE project. Conservation was calculated as the average phastCons value along each hit, as reported by modENCODE. Additional file 8. Accuracy evaluation of CBS predictions on modENCODE GAF/Trl ChIP-seq binding regions. At the top, the distribution of successfully identified ChIP-seq sites for CBS predictions is shown, taking into account different levels of sequence conservation. At the bottom, the ratio between the number of predictions and the number of successfully identified ChIP-seq sites for the same conservation thresholds is given. Additional file 9. Evaluation of CBS annotations in the even-skipped gene stripe 2 enhancer (GenBank: AF042709, dm3: chr2R:5865217–5865890). The following binding sites have been experimentally validated [78]: bicoid (+138, +159, +310, +403, +521), hunchback (+496, +578, +661), and Kruppel (+3, +139, +327, +521, +571, +615). At the top, we show the UCSC multiz15way conservation track. For each TF, we display the MatScan matrix score, CBS annotations, and experimental sites. This figure was graphically customized from original CBS results, incorporating the location of experimentally validated sites and the score of the weight matrix predictions into the final picture. (PDF 404 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Blanco, E., Corominas, M. CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes. BMC Genomics 13, 688 (2012). https://doi.org/10.1186/1471-2164-13-688
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-13-688