- Research
- Open access
- Published:
FunClust: a web server for the identification of structural motifs in a set of non-homologous protein structures
BMC Bioinformatics volume 9, Article number: S2 (2008)
Abstract
Background
The occurrence of very similar structural motifs brought about by different parts of non homologous proteins is often indicative of a common function. Indeed, relatively small local structures can mediate binding to a common partner, be it a protein, a nucleic acid, a cofactor or a substrate. While it is relatively easy to identify short amino acid or nucleotide sequence motifs in a given set of proteins or genes, and many methods do exist for this purpose, much more challenging is the identification of common local substructures, especially if they are formed by non consecutive residues in the sequence.
Results
Here we describe a publicly available tool, able to identify common structural motifs shared by different non homologous proteins in an unsupervised mode. The motifs can be as short as three residues and need not to be contiguous or even present in the same order in the sequence. Users can submit a set of protein structures deemed or not to share a common function (e.g. they bind similar ligands, or share a common epitope). The server finds and lists structural motifs composed of three or more spatially well conserved residues shared by at least three of the submitted structures. The method uses a local structural comparison algorithm to identify subsets of similar amino acids between each pair of input protein chains and a clustering procedure to group similarities shared among different structure pairs.
Conclusions
FunClust is fast, completely sequence independent, and does not need an a priori knowledge of the motif to be found. The output consists of a list of aligned structural matches displayed in both tabular and graphical form. We show here examples of its usefulness by searching for the largest common structural motifs in test sets of non homologous proteins and showing that the identified motifs correspond to a known common functional feature.
Background
More than a hundred methods have been developed so far for the automated discovery of unknown short conserved motifs in a set of protein or nucleic acid sequences [1]. These methods are routinely used for the identification of functional features, such as, for example, transcription factor binding sites in a set of gene regulatory regions.
However a functional motif needs not to be contiguous in sequence and might arise from the clustering in space of similar side chains coming from different parts of non homologous proteins. Finding occurrences of shared structural motifs can be instrumental for mapping the interaction site of different proteins with the same partner [2], for locating of the binding site for a common ligand even of unknown identity or for identifying an epitope shared, for example by an external agent and an endogenous protein involved in autoimmune diseases.
At present, several applications for the comparison of multiple structures are available [3, 4] and are used for clustering protein structures in families or for identifying large structural motifs shared by different folds. All these algorithms are based on fold comparison methods, and therefore the alignments they produce are often sequence-dependent and/or require at least a small core of conserved residues which are contiguous in the primary sequence [5, 6].
These fold comparison methods cannot be used to identify small structural motifs not conserved in sequence and belonging to non homologous proteins. Nevertheless identifying such motifs is important to study cases where the same metabolite, for example ATP, is bound to proteins with different folds [7, 8], or where the same protein interacts with different partners using the same surface [9].
Here we describe FunClust [10], a new web server for the identification of common structural motifs in a set of non homologous protein structures without any knowledge about the type or position of the motif, which, additionally, does not need to be present in all the submitted structures.
FunClust is based on a local (as opposed to global) structural comparison program [11]. Local structural comparison methods [12–15] can identify small sets of residues organized in a conserved geometry, irrespectively of the order in which they appear in the primary sequence. They are useful to search for functional and/or structural motifs such as active sites or ligand binding sites in non-homologous protein structures [16].
In this work we show that our method is able to effectively and efficiently identify common functional and/or structural motifs present in different structures. As test cases, we used proteins known to share common motifs and a complete set of ATP binding proteins. The server is also being used for finding common interface patches in proteins interacting with the same partner [2].
Results
The FunClust web server
The FunClust web server [10] enables the quick identification of structural motifs, putatively associated with a common function, present in an ensemble of non-homologous protein structures. Users submit a set of protein structures deemed to share a given function, binding capability or a common epitope, without the need to specify where the common structural motif should be located. The output consists of a list of one or more conserved sets of residues.
Input
The method accepts as input a set of PDB [17] codes or user-submitted coordinates in PDB file format. In the latter case, it is also possible to provide an arbitrary subset of residues instead of the whole protein. If a PDB code is given without any chain identifier, all the available chains are considered in the computation.
PDB codes and user-submitted coordinates are processed in order to validate their format and detect sequence similarities that can indicate homology relationships. Only one representative structure is used for each set of submitted protein chains with sequence identity higher than a user-specified threshold. This restriction reduces or eliminates the risk of finding local similarities arising because of the overall similarity among the input proteins.
The user can set the value of five parameters: the maximum r.m.s.d. among the involved residues, the maximum distance between the side chains of the residues forming the motif, the inclusion/exclusion of solvent accessible residues only, the inclusion/exclusion of hydrophobic residues and the weight of physicochemical similarities among the matched residues.
A set of pre-selected combinations of parameters tailored to the identification of four different kinds of functional sites (active sites, ligand binding sites, protein-protein interfaces and hydrophobic core packing motifs) is also provided.
Output
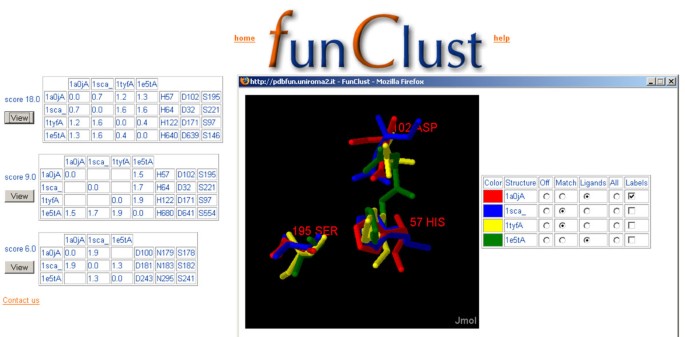
The output is a list of all the identified structural motifs, sorted by an approximate significance score, as described below. Each motif consists of a set of at least three protein chains and at least three residues in each chain. Not all the user-submitted structures need to contain the motif, but all the identified structures must contain a motif formed by the same number of residues. Each motif is shown in the output page along with its score in tabular and graphical form (figure 1). In the tabular form, the residues of each structure are listed in a different row, with the corresponding residues in different structures aligned in the same columns.
Details of structural matches. Output page of a FunClust search in four non-homologous serine protease protein chains (1a0jA, 1sca, 1tyfA and 1e5tA). The three different clusters identified are shown in tabular form. For each cluster the associated score is reported. The right section of each table reports, in each row, the residues belonging to the different structures, with structurally aligned residues written in the same column. The left section shows the r.m.s.d. value for each match identified between the structures corresponding to the row and column of the table (recall that two structures belonging to the cluster do not necessarily match to each other). In the example shown, the first of the three clusters is composed of the four catalytic triads which are therefore correctly identified. The second cluster identifies three non-catalytic residues in structure 1e5t, while the third one (with the lowest score) involves only three of the four structures. A user activated popup window shows a graphical view (created using the Jmol applet) of the first cluster. The four different structures have been superposed on the residues belonging to the structural motif. Each structure has a different colour, and only the residues involved in the cluster are shown. Commands to trigger the display of the whole structure and of the labels for each protein in the cluster are located in the right portion of the window.
Each motif is associated to its approximate significance score, which depends upon the number of protein chains containing it. More specifically, the score is given by the number of residues in the motif multiplied by the number of aligned pairs of structures. The score of a cluster formed by 3 aligned residues in 5 proteins chains can range from 3*10 (the maximum number of pairs formed by 5 structures) to 3*4 (the minimum number of pairs compatible with a motif identified in five structures).
Test cases
We show here some examples where the largest identified structural cluster of residues shared by a set of protein structures corresponds to a known functional motif. The examples are taken using PROSITE [18] regular expressions, CSA [19] catalytic residues, pdbFun residues in binding sites [20] and ELM [21] motifs. We selected our cases from the limited number of functional motifs identified on at least three structures with a completely different fold and low sequence identity. For each selected motif, we requested the representing structures to have a different CATH [22] architecture and share a sequence identity lower than 25%.
Some examples of cases where the first structural cluster identified by the server correctly pairs some or all of the correspondent functional residues in all the submitted structures are the serine endopeptidases enzymes (from CSA), the EF HAND motif (from ELM), the zinc binding site (from pdbFun) and the 4Fe-4S ferredoxin pattern (from PROSITE).
In Table 1 we show the results of the FunClust server for each different functional motifs. We also report the score and the set of parameters used (r.m.s.d. and side chain proximity).
ATP binding pockets
As an additional test of our method, we evaluated its ability to identify a common structural motif in a set of protein structures binding the same ligand. We used a complete set of 57 ATP binding structures [23] sharing less than 35% sequence identity, representative of all the PDB. For the comparison, all residues having at least one atom at less than 4.5Å from the bound ATP were used. Using the standard ligand binding site parameters of FunClust, a total of four structural motifs (cluster of residues) were identified. The three highest scoring clusters (96, 90 and 90) are composed respectively of 13, 16 and 19 different proteins all sharing a set of three residues: two Glycines and a Lysine or Serine. All residues in the three identified motifs belong to the p-loop binding motif [24]. By superposing the motifs, it can be seen that the vast majority of ATP molecules appear to have one of their phosphate atoms in the same location. The complete set of results can be viewed on the server web pages, using the “Example 3” set of structures.
Conclusions
Here we present a new server for the multiple local alignment of protein structures and show some examples of its application to the discovery of common functional patterns in serine endopeptidases, EF HAND containing proteins, ferredoxins, zinc and ATP binding proteins. FunClust is a useful tool in the automated discovery of local structural motifs shared by a set of non-homologous protein structures. The server is fast and easy to use. To date, this is the only method available on the web for the automated and unsupervised identification of local structural motifs in unrelated protein structures.
Methods
FunClust uses two different algorithms: a local structural comparison method that is able to identify all the similarities between a single pair of structures, and a procedure that searches for clusters of matches involving residues common to different structures. The result of the two procedures is a list of local structural motifs, each one identified by a cluster of structural matches between different pairs of structures.
Local comparison algorithm
In the first step of the procedure, all the input chains are compared pairwise using Query3D [11] a fast local comparison algorithm. This step involves, for n protein chains, n*(n-1)/2 comparisons. The method is able to find all the subsets of at least three residues that can be superposed within a given r.m.s.d. value and with sequence similarity above a user-defined threshold. The r.m.s.d is calculated using a two-point representation of each residue, comprising the C-alpha and the side-chain geometric centre.
Each comparison run generates a list of one or more sequence-independent local structural matches between the two proteins.
Clustering of structural matches
In the second step of the procedure a clustering algorithm identifies the largest structural matches shared by the highest number of structures.
To this end, a graph is built in which every node represents a match. Edges are drawn between nodes representing matches belonging to different lists (i. e. different pairs of structures involved in the comparison), according to the following criteria:
-
1.
the two lists must have one structure in common, i. e. they represent matches of the same structure with two different targets;
-
2.
the matches to be connected share at least three residues.
This graph is analyzed by a fast and simple procedure that searches for the largest number of connected nodes containing no more than one match from every list and at least three common residues in all the involved structures.
The algorithm selects the highest scoring set of connected matches in the graph. The score is given by the number of residues in common between all the matches multiplied by the number of matches belonging to the cluster.
When the highest scoring cluster is identified, the corresponding matches are removed from the graph and the search is repeated until all clusters are identified. Each cluster of matches corresponds to a different structural motif, with the score of the motif being that of the cluster.
Implementation notes
Both the comparison and clustering of multiple protein structures are complex problems, but extensive testing demonstrated that the CPU cost of our algorithm is fully compatible with protein structures of reasonable size and with motifs of average size present in up to 20 different structures. Comparison times range from fractions of a second to a few minutes. However, a time limit of 1 minute is given to web server users. Web pages have been tested using the most common browsers for Windows, Mac and Linux platforms.
References
Sandve GK, Drablos F: A survey of motif discovery methods in an integrated framework. Biol Direct 2006, 1: 11. 10.1186/1745-6150-1-11
Marcatili P., Bussotti G., Tramontano A.: The movin server for the analysis of protein interaction networks. BMC Bioinformatics 2008, 9(Suppl 2):S11. 10.1186/1471-2105-9-S2-S11
Novotny M, Madsen D, Kleywegt GJ: Evaluation of protein fold comparison servers. Proteins 2004, 54: 260–270. 10.1002/prot.10553
Hill AD, Reilly PJ: Comparing programs for rigid-body multiple structural superposition of proteins. Proteins 2006, 64: 219–226. 10.1002/prot.20975
Shatsky M, Shulman-Peleg A, Nussinov R, Wolfson HJ: The multiple common point set problem and its application to molecule binding pattern detection. J Comput Biol 2006, 13: 407–428. 10.1089/cmb.2006.13.407
Shatsky M, Nussinov R, Wolfson HJ: A method for simultaneous alignment of multiple protein structures. Proteins 2004, 56: 143–156. 10.1002/prot.10628
Moodie SL, Mitchell JB, Thornton JM: Protein recognition of adenylate: an example of a fuzzy recognition template. J Mol Biol 1996, 263: 486–500. 10.1006/jmbi.1996.0591
Denessiouk KA, Rantanen VV, Johnson MS: Adenine recognition: a motif present in atp-, coa-, nad-, nadp-, and fad-dependent proteins. Proteins 2001, 44: 282–291. 10.1002/prot.1093
Henschel A, Kim WK, Schroeder M: Equivalent binding sites reveal convergently evolved interaction motifs. Bioinformatics 2006, 22: 550–555. 10.1093/bioinformatics/bti782
The FunClust web server[http://pdbfun.uniroma2.it/funclust]
Ausiello G, Via A, Helmer-Citterich M: Query3d: a new method for high-throughput analysis of functional residues in protein structures. BMC Bioinformatics 2005, 6(Suppl 4):S5. 10.1186/1471-2105-6-S4-S5
Wallace AC, Laskowski RA, Thornton JM: Derivation of 3d coordinate templates for searching structural databases: application to ser-his-asp catalytic triads in the serine proteinases and lipases. Protein Sci 1996, 5: 1001–1013.
Stark A, Sunyaev S, Russell RB: A model for statistical significance of local similarities in structure. J Mol Biol 2003, 326: 1307–1316. 10.1016/S0022-2836(03)00045-7
Jones S, Thornton JM: Searching for functional sites in protein structures. Curr Opin Chem Biol 2004, 8: 3–7. 10.1016/j.cbpa.2003.11.001
Ferre F, Ausiello G, Zanzoni A, Helmer-Citterich M: Functional annotation by identification of local surface similarities: a novel tool for structural genomics. BMC Bioinformatics 2005, 6: 194. 10.1186/1471-2105-6-194
Gherardini PF, Wass MN, Helmer-Citterich M, Sternberg MJE: Convergent evolution of enzyme active sites is not a rare phenomenon. J Mol Biol 2007, 372: 817–845. 10.1016/j.jmb.2007.06.017
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The protein data bank. Nucleic Acids Res 2000, 28: 235–242. 10.1093/nar/28.1.235
Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA: The prosite database. Nucleic Acids Res 2006, 34: D227–30. 10.1093/nar/gkj063
Porter CT, Bartlett GJ, Thornton JM: The catalytic site atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res 2004, 32: D129–33. 10.1093/nar/gkh028
Ausiello G, Zanzoni A, Peluso D, Via A, Helmer-Citterich M: Pdbfun: mass selection and fast comparison of annotated pdb residues. Nucleic Acids Res 2005, 33: W133–7. 10.1093/nar/gki499
Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ: Elm server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res 2003, 31: 3625–3630. 10.1093/nar/gkg545
Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, Sillitoe I, Yeats C, Thornton JM, Orengo CA: The cath domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res 2007, 35: D291–7. 10.1093/nar/gkl959
Stockwell GR, Thornton JM: Conformational diversity of ligands bound to proteins. J Mol Biol 2006, 356: 928–944. 10.1016/j.jmb.2005.12.012
Via A, Ferre F, Brannetti B, Valencia A, Helmer-Citterich M: Three-dimensional view of the surface motif associated with the p-loop structure: cis and trans cases of convergent evolution. J Mol Biol 2000, 303: 455–465. 10.1006/jmbi.2000.4151
Acknowledgements
We thank Luca Parca for his useful contribution. This work was supported by Telethon (GGP04273), AIRC, and FIRB projects LIBI and ITALBIONET.
This article has been published as part of BMC Bioinformatics Volume 9 Supplement 2, 2008: Italian Society of Bioinformatics (BITS): Annual Meeting 2007. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2105/9?issue=S2
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GA conceived the study, carried out the work and drafted the manuscript. PM helped in testing the server. PFG, AV and AT participated in the design of the work. MHC participated in the design of the work and in its coordination. All authors read and approved the final manuscript.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ausiello, G., Gherardini, P.F., Marcatili, P. et al. FunClust: a web server for the identification of structural motifs in a set of non-homologous protein structures. BMC Bioinformatics 9 (Suppl 2), S2 (2008). https://doi.org/10.1186/1471-2105-9-S2-S2
Published:
DOI: https://doi.org/10.1186/1471-2105-9-S2-S2